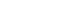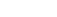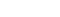Background and Purpose
Over sixty million people worldwide are estimated to sustain a traumatic brain injury (TBI) each year. Mild TBI (mTBI) accounts for over 80% of all individuals with TBI. In spite of the generally good prognosis, symptoms following mTBI can persist for more than a year, including an inability to concentrate, memory deficits, reduced problem-solving capacity, and impaired balance. There are no well-validated diagnostic tools to predict which patients are likely to experience sustaining problems, which is called post-concussion syndrome (PCS). Furthermore, TBI increases a risk for degenerative neurological diseases in the later life. However, the mechanisms contributing to the development of PCS or neurodegeneration after TBI still remain to be elucidated. The present study aimed to investigate the TBI-induced molecular dysfunctions in the mouse brain using the proteomics approach.
Materials and Methods
In this study, quantitative proteomic approach, a high-resolution Q-exactive mass spectrometer, was employed to measure proteome changes of mouse brain tissue after mTBI. A total of six 2-month-old male C57BL6 mice were randomly assigned to either the control (n=3) or the weight-drop mTBI group (n=3). The mouse in mTBI group was mounted on a thin foil and was impacted on the mid-point between the bregma and lambda by a single metallic weight. The whole brain tissues were harvested 72 hours after the injury and were pooled for the subsequent proteomic analysis.
Results
A total of 3 047 proteins were identified in >90% of the samples, of which, 432 proteins were significantly changed (differentially expressed proteins, DEPs) between control and mTBI groups. When the cut-off value 1.5 for the fold change was applied, 250 proteins were yielded (65 up- and 185 down-regulated proteins, Figure 1). Functional bioinformatics analysis and protein to protein interaction (PPI) network mapping showed biological processes such as the oxidative reduction, protein translation and RNA processing, protein transport, and glial cell differentiation were over-represented. Ingenuity pathway analysis (IPA) revealed that the most affected biological function by the DEPs was the long-term potentiation (Figure 2).
Conclusion
These results demonstrated that mTBI evokes distinct proteome changes after the injury, and the long-term potentiation become a most vulnerable biological function to the following pathophysiologic cascade.
|
|
Fig 1. Included, detected, and differentially-expressed proteins
Fig 2. A cluster analysis for the differentially expressed proteins with the fold change >1.5
Fig 3. Protein network analysis using Ingenuity Pathway Analysis. Long-term potentiation was shown to be the most affected function by the proteome changes after mTBI.
|